Genetic Variation and HCV Genotyping
Even in a single infected individual, HCV does not exist as a homogeneous species. Heterogeneous genomes – “quasispecies” – resulting from mutations due to high error rates in RNA replication are found within the same host. Many important biological features of several viruses are attributable to their quasispecies nature, including vaccination failure, persistent infection, and resistance to antiviral drugs.38 The amount of diversity of the quasispecies population has also been found to be related to the progression to liver disease.
The most striking feature of HCV is its ability to persist in the host. The mechanism(s) of viral persistence are unclear but cannot include viral integration into the host genome as with certain other viruses due to the lack of a DNA intermediate in its life cycle. Instead, persistence appears to result from HCV’s ability to mutate rapidly under immune pressure, giving rise to related but immunologically distinct variants.11 Any one of these variants can become the predominant strain, and coexistence of multiple quasispecies allows HCV to escape the host immune response.
Most mutations occur in a short, hypervariable region of the E2/NS1 domain. The region represents only 8% of the domain but accounts for approximately half of the nucleotide changes in the entire envelope region.39 Because of the occurrence of this hypervariable region within the envelope where it would be most likely to be exposed to antibody, mutations in this region may serve to evade an immune response.11,39 It has been reported that the nucleotide substitution rate within the hypervariable region rises during acute infection at a time when HCV RNA levels in the serum are decreasing, possibly due to a host immune response.40 There is also evidence that HCV can escape immune clearance by down-regulating its replication while persisting quiescently in the liver.11
Genotypic analysis and prevalence
Traditionally, viruses have been classified according to antigenic characteristics, but with recent advances in molecular biology, genotypic classification through the analysis of genomic variation is now possible. Variations in the HCV genome fall into a series of specific patterns that have been classified into genotypes.41 Among the different HCV genotypes, the sequence of the 5′ NC region is relatively conserved and is most often applied for diagnosis of HCV infection by PCR. In contrast, the sequences of NS3, NS5, and core regions are more variable and are therefore often used to define and distinguish among the HCV genotypes.42
Studies indicate that there are nine major HCV types (according to the general classification) designated 1 through 9.18
Some of these types are further divided into subtypes. The potential significance of this becomes apparent when considering virus-host interactions, severity of infection, and sensitivity to treatment. The clinical importance of HCV lies in its persistence and ability to cause chronic liver disease. The dramatic disparities in HCV disease course among infected persons and differences in disease patterns between countries with divergent dominant genotypes raise the possibility that the existence of various strains of HCV may be a critical factor in this variability.
In one study, 98 patients in the United States with chronic Hepatitis C infection were examined (Fig 1).41 Type 1 was the dominant genotype, present in 74% of patients, almost evenly divided between subtypes 1a and 1b. Two additional patients had a dual infection with types 1 and 2 while another case had both subtypes 1a and 1b.
One patient was untypeable. There was no correlation between infecting genotype and presumed cause, serum indices of necroinflammatory activity, age or sex of the patients, or known duration of infection. Patients with HCV genotype 2 had more severe liver disease histologically than did patients with other genotypes but paradoxically had significantly lower circulating levels of Hepatitis C viral RNA. 41 A second, independent study, also performed in the United States, found that HCV genotype 1 represented 70% of viral isolates, confirming that it accounts for the majority of infections in American patients. 43
Fig 1. — Prevalence of HCV genotypes and subtypes among American patients N=98).41
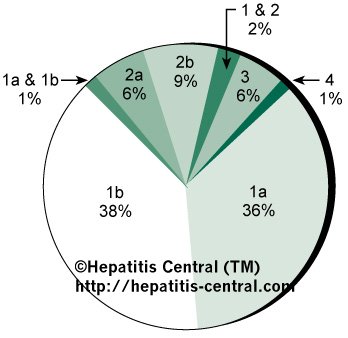
Segments in which more than one genotype or subtype appears represent mixed infections.
Another study performed in Japan (using study-specific genotype nomenclature) demonstrated striking differences in the distribution of HCV genotypes in various countries.44 This group classified HCV into four main types based on variations in the NS5 region and studied the prevalence of these types in Japan, China, Europe, Brazil, and the United States. Results revealed that the prevalence of one particular strain was the highest in every country except the United States, making it the probable major type of HCV worldwide. Another strain was rare in Japanese and Chinese samples but common in samples from Western countries, suggesting the existence of a Western HCV type. Similarly, a third strain was rare in Western samples but common in Japanese and Chinese samples, suggesting the existence of an Eastern HCV type. Because of the study-specific classification utilized, however, comparisons to the results of other studies are not possible.
Despite reports of an interplay between genotype and disease severity, patients with the same genotype can have very different clinical outcomes. Thus, differences in genotype may represent only part of a complex interaction between host and virus. In addition, the effect of genotypes may be indirect in that they may reflect differences in viral burden rather than differences in strain virulence.11 There is, however, evidence for genotypic variability in the response of HCV to therapy.
Serotyping
Serotype-specific immunodominant epitopes have recently been mapped to the NS4 region of HCV. 45 Synthetic peptides from this region can be used as capture antigens in an ELISA assay to differentiate serologically among infections with the various types of HCV. Blocking antibody or competing peptide is used to overcome cross-reactivity. The results of genotypic and serologic analyses are well correlated for genotypes and serotypes 1, 2, and 3, which are predominant in the U.S. population.46 The value of serotyping relative to genotyping remains to be determined.
References
11. Alter HJ. To C or not to C: these are the questions. Blood. 1995;85:1681-1695.
18. Berenguer M, Wright TL. Hepatitis C virus. Advances in Gastroenterology, Hepatology & Clinical Nutrition. 1996;1:2-21.
38. Kanazawa Y, Hayashi N, Mita E, et al. Influence of viral quasispecies on effectiveness of interferon therapy in chronic Hepatitis C patients. Hepatology. 1994;20:1121-1130.
39. Weiner AJ, Brauer MJ, Rosenblatt J, et al. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842-848.
40. Yamaguchi K, Tanaka E, Higashi K, et al. Adaptation of Hepatitis C virus for persistent infection in patients with acute hepatitis. Gastroenterology. 1994;106:1344-1248.
41. Mahaney K, Tedeschi V, Maertens G, et al. Genotypic analysis of Hepatitis C virus in American patients. Hepatology. 1994;20:1405-1411.
42. Qu D, Li J-S, Vitvitski L, et al. Hepatitis C virus genotypes in France: comparison of clinical features of patients infected with HCV type I and type II. J Hepatol. 1994;21:70-75.
43. Lau JYN, Davis GL, Ohno T, et al. Application of Hepatitis C virus (HCV) subtyping in chronic Hepatitis C in the United States. Hepatology. 1993;18:149A. Abstract.
44. Takada N, Takase S, Takada A, et al. Differences in the Hepatitis C virus genotypes in different countries. J Hepatol. 1993;17:277-283.
45. Chemello L, Alberti A, Rose K, et al. Hepatitis C serotype and response to interferon therapy. N Engl J Med. 1994; 330:143-144. Correspondence.
46. Keefe E, Blatt LM, Dusheiko G, et al. Differential response to treatment with consensus interferon (CIFN) and IFN a-2b in chronic HCV patients infected with different HCV serotypes. Hepatology. 1996;24:275A. Abstract.




